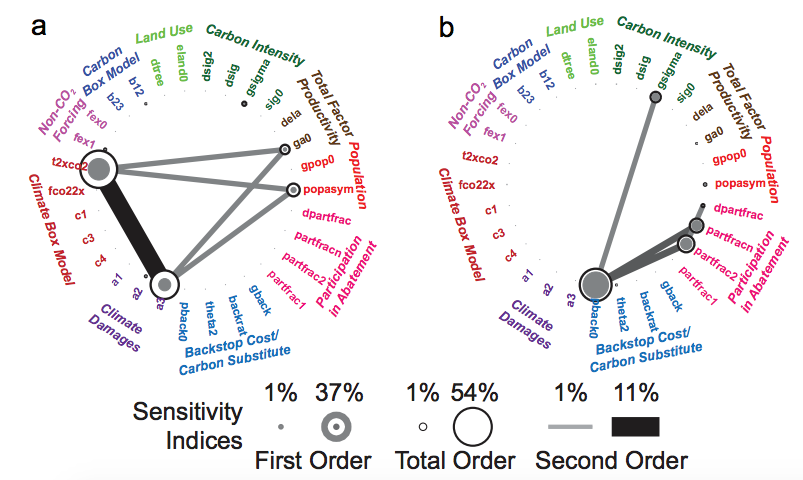
Sensitivity analysis aims at assessing the relative contributions of the different sources of uncertainty to the variability in the output of a model. There are several mathematical techniques available in the literature, with variance-based approaches being the most popular, and variance-based indices the most widely used, especially “total effects” indices. Literature also provides several estimators/approximators for these indices (reviews can be found here [1]), which typically need N = n × (M + 2) model evaluations (and resulting outputs), where M is the number of uncertain inputs and n is some factor of proportionality that is selected ad hoc, depending on the complexity of the model (e.g. linear or not, monotonic or not, continuous or not). [Note: Literature typically refers to n as the number of samples and to N as the number of model evaluations, and this is how I’ll be using them also.]
The generation of this set of model runs of size N is by far the most computationally demanding step in the calculation of variance-based indices, and a major limitation to their applicability, as it’s typically in the order of magnitude of thousands [2] (n typically >1000). For example, consider a model with 5 uncertain inputs that takes 1 minute to run. To appropriately estimate sensitivity indices for such a model, we would potentially need about N=7000 model evaluations, which would take almost 5 days on a personal computer, excluding the time for the estimator calculation itself (which is typically very short).
The aim is therefore to pick the minimum n needed to ensure our index calculation is reliable. Unfortunately, there is no hard and fast rule on how to do that, but the aim of this post is to illuminate that question a little bit and provide some guidance. I am going off the work presented here [3] and elsewhere, and the aim is to perform the estimation of sensitivity indices repeatedly, using an increasing number of n until the index values converge.
I will be doing this using a fishery model, which is a nonlinear and nonmonotonic model with 9 parameters. Based on previous results suggesting that 3 of these parameters are largely inconsequential to the variability in the output, I’ll be fixing them to their nominal values. I’ll be using SALib to perform the analysis. My full script can be found here, but I’ll only be posting the most important snippets of code in this post.
First, I set up my SALib ‘problem’ and create arrays to store my results:
# Set up dictionary with system parameters
problem = {
'num_vars': 6,
'names': ['a', 'b', 'c','h',
'K','m'],
'bounds': [[ 0.002, 2],
[0.005, 1],
[0.2, 1],
[0.001, 1],
[100, 5000],
[0.1, 1.5]]
}
# Array with n's to use
nsamples = np.arange(50, 4050, 50)
# Arrays to store the index estimates
S1_estimates = np.zeros([problem['num_vars'],len(nsamples)])
ST_estimates = np.zeros([problem['num_vars'],len(nsamples)])
I then loop through all possible n values and perform the sensitivity analysis:
# Loop through all n values, create sample, evaluate model and estimate S1 & ST
for i in range(len(nsamples)):
print('n= '+ str(nsamples[i]))
# Generate samples
sampleset = saltelli.sample(problem, nsamples[i],calc_second_order=False)
# Run model for all samples
output = [fish_game(*sampleset[j,:]) for j in range(len(sampleset))]
# Perform analysis
results = sobol.analyze(problem, np.asarray(output), calc_second_order=False,print_to_console=False)
# Store estimates
ST_estimates[:,i]=results['ST']
S1_estimates[:,i]=results['S1']
I can then plot the evolution of these estimates as n increases:

What these figures tell us is that choosing an n below 1000 for this model would potentially misestimate our indices, especially the first order ones (S1). As n increases, we see the estimates becoming less noisy and converging to their values. For more complex models, say, with more interactive effects, the minimum n before convergence could actually be a lot higher. A similar experiment by Nossent et al. [3] found that convergence was reached only after n=12,000.
An observation here is that the values of the total indices (ST) are higher than those of the the first order indices (S1), which makes sense, as ST includes both first order effects (captured by S1) and second order effects (i.e. interactions between the factors). Another observation here is that the parameters with the most significant effects (m and K) converge much faster than parameters with less impact on the output (a and b). This was also observed by Nossent et al. [3].
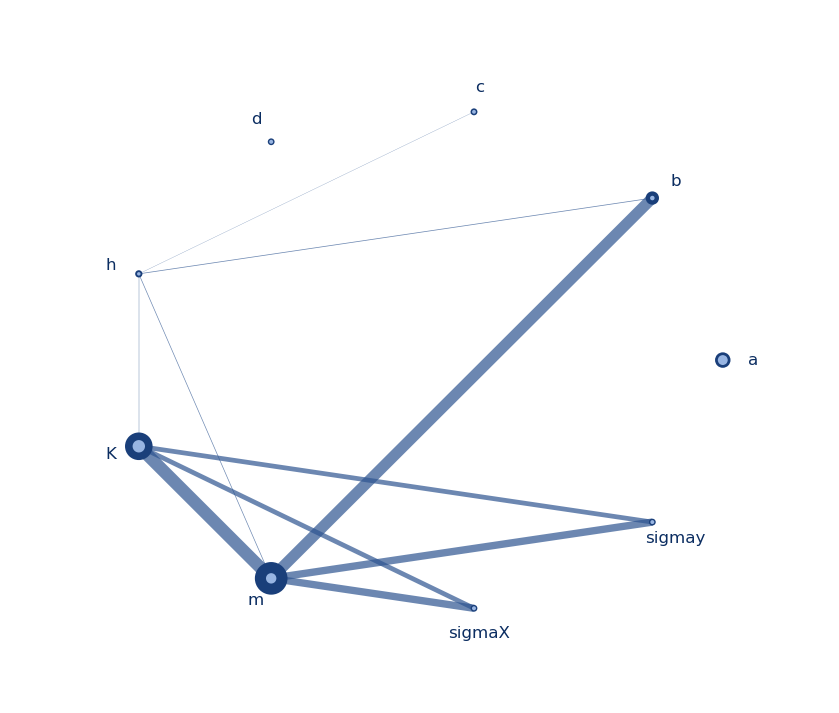
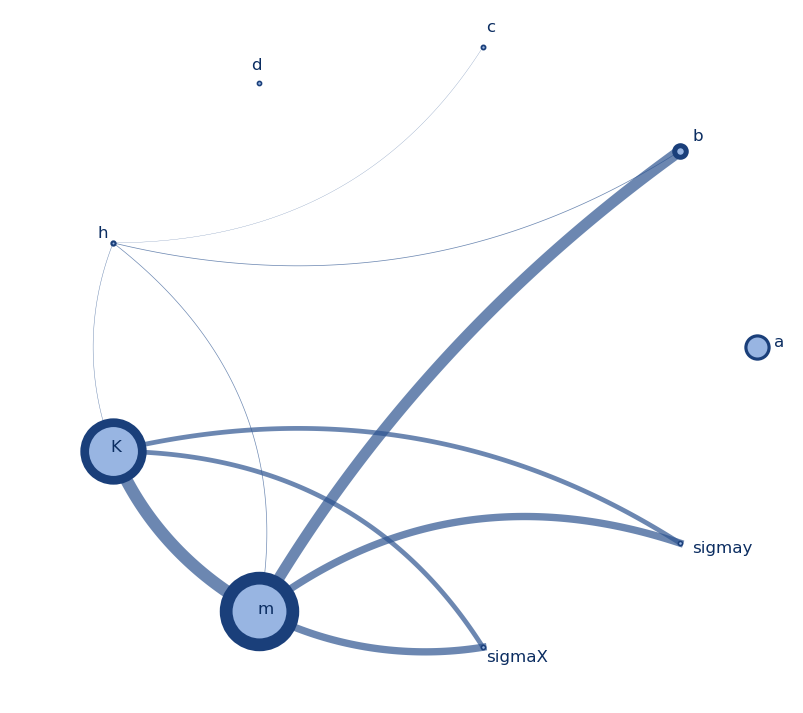
Finally, sensitivity analysis is often performed for the purposes of factor prioritization, i.e. determining (often rank-ordering) the most important parameters for the purposes of, for example, deciding where to devote most calibration efforts in the model or most further analysis to reduce the uncertainty in a parameter. The figures below show the evolution of that rank-ordering as we increase n.

These figures show that with a number of samples that is too small, we could potentially misclassify a factor as important or unimportant when it actually is not.
Now, one might ask: how is this useful? I’m trying to minimize my n, but you’ve just made me run way too many model evaluations, multiple times, just to determine how I could have done it faster? Isn’t that backwards?
Well, yes and no. I’ve devoted this time to run this bigger experiment to get insight on the behavior of my model. I have established confidence in my index values and factor prioritization. Further, I now know that n>1500 would probably be unnecessary for this system and even if the model itself or my parameter ranges change. As long as the parameter interactions, and model complexity remain relatively the same, I can leverage this information to perform future sensitivity analyses, with a known minimum n needed.
References:
[1] A. Saltelli, P. Annoni, I. Azzini, F. Campolongo, M. Ratto, and S. Tarantola, “Variance based sensitivity analysis of model output. Design and estimator for the total sensitivity index,” Computer Physics Communications, vol. 181, no. 2, pp. 259–270, Feb. 2010, doi: 10.1016/j.cpc.2009.09.018.
[2] F. Pianosi and T. Wagener, “A simple and efficient method for global sensitivity analysis based on cumulative distribution functions,” Environmental Modelling & Software, vol. 67, pp. 1–11, May 2015, doi: 10.1016/j.envsoft.2015.01.004.
[3] J. Nossent, P. Elsen, and W. Bauwens, “Sobol’ sensitivity analysis of a complex environmental model,” Environmental Modelling & Software, vol. 26, no. 12, pp. 1515–1525, Dec. 2011, doi: 10.1016/j.envsoft.2011.08.010.