As most of you probably know, scenario discovery is an exploratory modeling approach [Bankes, 1993] that involves stress-testing proposed policies over plausible future “states of the world” (SOWs) to discover conditions under which those policies would fail to meet performance goals [Bryant and Lempert, 2010]. The scenario discovery process is therefore an exercise in statistical classification. Two commonly used methods used for the scenario discovery process are the Patient Rule Induction Method (PRIM; Friedman and Fisher [1999]) and Classification and Regression Trees (CART; Breiman et al. [1984]), both of which are included in the OpenMORDM R package and Rhodium Python package.
Another commonly used method in classification that hasn’t been given much attention in the scenario discovery literature is logistic regression. Logistic regression models estimate the probability that an event is classified as a success (1) as opposed to a failure (0) as a function of different covariates. This allows for the definition of “safe operating spaces,” or factor combinations leading to success, based on the probability with which one would like to be able to achieve the specified performance goal(s). We may not know the probability that a particular SOW will occur, but through the logistic regression we can estimate the probability of success in that SOW should it occur. The logistic regression can also identify which factors most influence a policy’s ability to meet those performance goals.
This blog post will illustrate how to build logistic regression models in Python for scenario discovery using the Red River basin as an example. Here we are interested in determining under what streamflow and demand characteristics reservoir operating policies are unable to protect Hanoi from the 100-yr flood. We assume operators want to ensure protection to this event with at least 95% reliability and use logistic regression to estimate under what combination of streamflow and demand characteristics they will be able to do so.
The form of the logistic regression model is given by Equation 1, where pi represents the probability that performance in the ith SOW is classified as a success and Xi represents a vector of covariates (in this case, streamflow and demand characteristics) describing the ith SOW:
1)  .
.
The coefficients,  , on the covariates are estimated using Maximum Likelihood Estimation.
, on the covariates are estimated using Maximum Likelihood Estimation.
To determine which streamflow and demand characteristics are most important in explaining successes and failures, we can compare the McFadden’s pseudo-R2 values associated with different models that include different covariates. McFadden’s pseudo-R2,  , is given by Equation 2:
, is given by Equation 2:
2) 
where 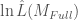 is the log-likelihood of the full model and
is the log-likelihood of the full model and  is the log-likelihood of the intercept model, i.e. a model with no covariates beyond the intercept. The intercept model therefore predicts the mean probability of success across all SOWs.
is the log-likelihood of the intercept model, i.e. a model with no covariates beyond the intercept. The intercept model therefore predicts the mean probability of success across all SOWs.  is a measure of improvement of the full model over the intercept model.
is a measure of improvement of the full model over the intercept model.
A common approach to fitting regression models is to add covariates one-by-one based on which most increase R2 (or in this case,  ), stopping once the increase of an additional covariate is marginal. The covariate that by itself most increases
), stopping once the increase of an additional covariate is marginal. The covariate that by itself most increases  is therefore the most important in predicting a policy’s success. To do this in Python, we will use the library
is therefore the most important in predicting a policy’s success. To do this in Python, we will use the library statsmodels.
Imagine we have a pandas dataframe, dta that includes n columns of streamflow and demand characteristics describing different SOWs (rows) and a final column of 0s and 1s representing whether or not the policy being evaluated can provide protection to the 100-yr flood in that SOW (0 for no and 1 for yes). Assume the column of 0s and 1s is the last column and it is labeled Success. We can find the value of  for each covariate individually by running the following code:
for each covariate individually by running the following code:
import pandas as pd
import statsmodels.api as sm
from scipy import stats
# deal with fact that calling result.summary() in statsmodels.api
# calls scipy.stats.chisqprob, which no longer exists
stats.chisqprob = lambda chisq, df: stats.chi2.sf(chisq, df)
def fitLogit(dta, predictors):
# concatenate intercept column of 1s
dta['Intercept'] = np.ones(np.shape(dta)[0])
# get columns of predictors
cols = dta.columns.tolist()[-1:] + predictors
#fit logistic regression
logit = sm.Logit(dta['Success'], dta[cols])
result = logit.fit()
return result
dta = pd.read_csv('SampleData.txt')
n = len(dta.columns) - 1
for i in range(n):
predictors = dta.columns.tolist()[i:(i+1)]
result = fitLogit(dta, predictors)
print(result.summary())
A sample output for one predictor, Col1 is shown below. This predictor has a pseudo-R2 of 0.1138.

Once the most informative predictor has been determined, additional models can be tested by adding more predictors one-by-one as described above. Suppose that through this process, one finds that the first 3 columns of dta (Col1,Col2 and Col3) are the most informative for predicting success on providing protection to the 100-yr flood, while the subsequent columns provide little additional predictive power. We can use this model to visualize the probability of success as a function of these 3 factors using a contour map. If we want to show this as a 2D projection, the probability of success can only be shown for combinations of 2 of these factors. In this case, we can hold the third factor constant at some value, say its base value. This is illustrated in the code below, which also shows a scatter plot of the SOWs. The dots are shaded light blue if the policy succeeds in providing protection to the 100-yr flood in that world, and dark red if it does not.
import numpy as np
import matplotlib.pyplot as plt
import matplotlib as mpl
import seaborn as sns
import pandas as pd
import statsmodels.api as sm
def fitLogit(dta, predictors):
# concatenate intercept column of 1s
dta['Intercept'] = np.ones(np.shape(dta)[0])
# get columns of predictors
cols = dta.columns.tolist()[-1:] + predictors
#fit logistic regression
logit = sm.Logit(dta['Success'], dta[cols])
result = logit.fit()
return result
def plotContourMap(ax, result, constant, dta, contour_cmap, dot_cmap, levels, xgrid, ygrid, \
xvar, yvar, base):
# find probability of success for x=xgrid, y=ygrid
X, Y = np.meshgrid(xgrid, ygrid)
x = X.flatten()
y = Y.flatten()
if constant == 'x3': # 3rd predictor held constant at base value
grid = np.column_stack([np.ones(len(x)),x,y,np.ones(len(x))*base[2]])
elif constant == 'x2': # 2nd predictor held constant at base value
grid = np.column_stack([np.ones(len(x)),x,np.ones(len(x))*base[1],y])
else: # 1st predictor held constant at base value
grid = np.column_stack([np.ones(len(x)),np.ones(len(x))*base[0],x,y])
z = result.predict(grid)
Z = np.reshape(z, np.shape(X))
contourset = ax.contourf(X, Y, Z, levels, cmap=contour_cmap)
ax.scatter(dta[xvar].values, dta[yvar].values, c=dta['Success'].values, edgecolor='none', cmap=dot_cmap)
ax.set_xlim(np.min(X),np.max(X))
ax.set_ylim(np.min(Y),np.max(Y))
ax.set_xlabel(xvar,fontsize=24)
ax.set_ylabel(yvar,fontsize=24)
ax.tick_params(axis='both',labelsize=18)
return contourset
# build logistic regression model with first 3 columns of predictors from dta
dta = pd.read_csv('SampleData.txt')
predictors = dta.columns.tolist()[0:3]
result = fitLogit(dta, predictors)
# define color map for dots representing SOWs in which the policy
# succeeds (light blue) and fails (dark red)
dot_cmap = mpl.colors.ListedColormap(np.array([[227,26,28],[166,206,227]])/255.0)
# define color map for probability contours
contour_cmap = mpl.cm.get_cmap(‘RdBu’)
# define probability contours
contour_levels = np.arange(0.0, 1.05,0.1)
# define grid of x (1st predictor), y (2nd predictor), and z (3rd predictor) dimensions
# to plot contour map over
xgrid = np.arange(-0.1,1.1,0.01)
ygrid = np.arange(-0.1,1.1,0.01)
zgrid = np.arange(-0.1,1.1,0.01)
# define base values of 3 predictors
base = [0.5, 0.5, 0.5]
fig = plt.figure()
ax = fig.add_subplot(121)
# plot contour map when 3rd predictor ('x3') is held constant
plotContourMap(ax, result, 'x3', dta, contour_cmap, dot_cmap, contour_levels, xgrid, ygrid, \
'Col1', 'Col2', base)
ax = fig.add_subplot(122)
# plot contour map when 2nd predictor ('x2') is held constant
contourset = plotContourMap(ax, result, 'x2', dta, contour_cmap, dot_cmap, contour_levels, xgrid, zgrid, \
'Col1', 'Col3', base)
fig.subplots_adjust(wspace=0.3,hspace=0.3,right=0.8)
cbar_ax = fig.add_axes([0.85, 0.15, 0.05, 0.7])
cbar = fig.colorbar(contourset, cax=cbar_ax)
cbar_ax.set_ylabel('Probability of Success',fontsize=20)
yticklabels = cbar.ax.get_yticklabels()
cbar.ax.set_yticklabels(yticklabels,fontsize=18)
fig.set_size_inches([14.5,8])
fig.savefig('Fig1.png')
fig.clf()
This produces the following figure:

We can also use the probability contours discovered above to define “safe operating spaces” as combinations of these 3 factors under which the evaluated policy is able to succeed in providing protection to the 100-yr flood with some reliability, say 95%. The hyperplane of factor combinations defining that 95% probability contour can be determined by setting p to 0.95 in Equation 2. Again, to plot 2-D projections of that hyperplane, the values of the other covariates can be held constant at their base values. The code below illustrates how to do this with a 95% boundary.
# define colormap for classifying boundary between failure and success
class_cmap = mpl.colors.ListedColormap(np.array([[251,154,153],[31,120,180]])/255.0)
# define probability cutoff between failure and success
class_levels = [0.0, 0.95, 1.0]
fig = plt.figure()
ax = fig.add_subplot(121)
# plot contour map when 3rd predictor ('x3') is held constant
plotContourMap(ax, result, 'x3', dta, class_cmap, dot_cmap, class_levels, xgrid, ygrid, \
'Col1', 'Col2', base)
ax = fig.add_subplot(122)
# plot contour map when 2nd predictor ('x2') is held constant
plotContourMap(ax, result, 'x2', dta, class_cmap, dot_cmap, class_levels, xgrid, zgrid, \
'Col1', 'Col3', base)
fig.set_size_inches([14.5,8])
fig.savefig('Fig2.png')
fig.clf()
This produces the following figure, where the light red region is the parameter ranges in which the policy cannot provide protection to the 100-yr flood with 95% reliability, and the dark blue region is the “safe operating space” in which it can.

All code for this example can be found here.










